
Cedar Crest - Touch on Different Fabrics
Objective: This research focused on the M-Vac™ System's ability to collect low template or touch DNA from fabric.
Background and Methods: Low template DNA (LT-DNA), also known as low-copy-number DNA, trace or touch, is defined as a sample that contains less than 100 pg of DNA and it is often found at a crime scene in the form of finger or hand prints.
Traditional sampling techniques used to collect biological samples are swabbing, taping and cutting. A common issue with these methods and LT-DNA is that they cannot always collect enough DNA to get a full profile. The M-Vac™ has a significantly higher recovery rates and is projected to be a great addition to traditional techniques.
One of the questions about this new technology is where to use the M-Vac™. In this research, fabrics are evaluated relative to touch DNA. The following ten substrates were included in the evaluation – Knitted Cotton, Woven Cotton, Gabardine Polyester, Silk Polyester, Nylon, Rayon, Wool, Linen, 95% Cotton/5% Spandex and Acetate. The test swatches were 5 cm x 10 cm and handled for 10 seconds. Three different participants handled each type of fabric 5 times for a total of 15 replicates per fabric type.
Collections were done by one serologist using approximately 100-125 mL of surface rinse solution per collection. The collected samples were concentrated using 0.45 micrometer PES filters. The filters were cut into small pieces, placed in a microcentrifuge tube and extracted using Promega DNA IQTM Extraction, quantified with qPCR using Alu primers and SYBR Green, amplified with Promega PowerPlex® 16 Amplification and analyzed with ABI PRISM® 310 and GeneMapper IDX software. Only collections with DNA yields of more than 20 picograms were amplified and analyzed.
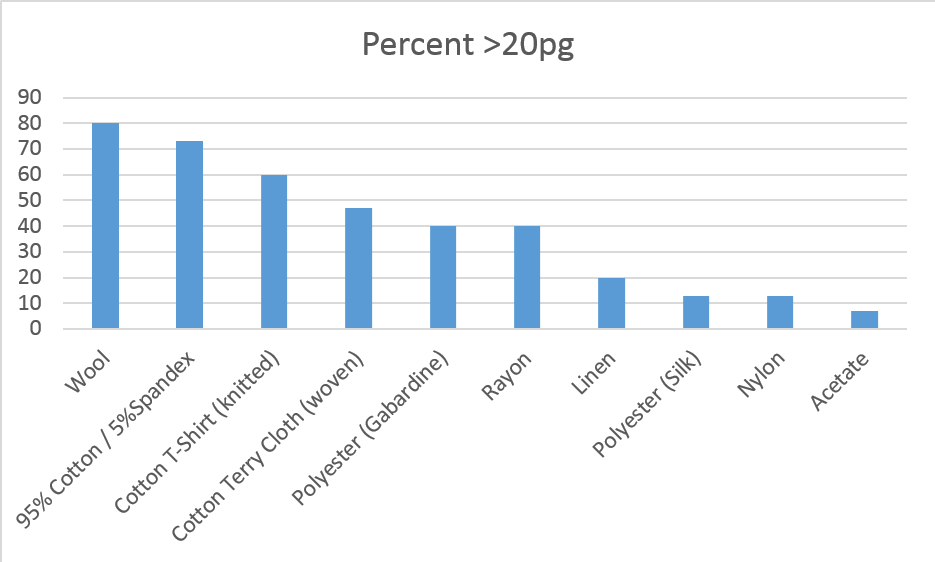
Results: Wool, 95% Cotton/5% Spandex and knitted cotton yielded statistically more DNA material than the other fabrics. 80 percent of the wool, 73 percent of the 95% Cotton/5% Spandex and 60 percent of the knitted cotton yielded more than the 20 pg threshold minimum for amplification. The chart below shows the yield percentages by fabric.
From literature, 5-6 percent of LT-DNA samples yield full profiles. The M-Vac™ collections yielded 49% of the samples amplified and 19% of the samples that were taken. 59 of the 150 samples taken yielded more than 20 pg of DNA.
From literature, 17-20 percent of LT-DNA samples yield profiles that can be loaded into CODIS. The M-Vac™ collections yielded CODIS enterable profiles from 75% of the samples amplified.
Since no direct comparisons were made as part of the research with traditional methods, only historical or literature comparisons could be made.
Conclusions:
- 39% of the collections had more than 20 pg of DNA
- 75% of the amplified profiles were suitable for CODIS
Without direct comparison testing, it is difficult to know what the exact comparison would be. However, the simple comparison with historical results was positive. Further research may consider doing direct comparisons with traditional collection methods, amplifications and analysis without the cut off threshold of 20 pg and larger swatches of material.
Acknowledgments: This research was conducted by Danielle Weinstein and Janine Kishbaugh of Cedar Crest as part of a thesis. It was presented at the 2014 NEAFS Conference. The presentation is available upon request.
Technical Note: The M-Vac™ collection technique used in the experiment was a side to side motion instead of the recommended forward and back motion. This may have negatively affected the results due to the fact that the spray comes out of the nozzle in a fan shape. A side to side motion would only impact the surface with a line of solution rather than the full width of the fan with a forward and back motion.