
Insights on Pre-filtering
 University of California-Davis student Vianca Castro-Harris studied several aspects of the M-Vac™ System use in her masters program project, including the efficacy of pre-filtering. Most of the previous studies she reviewed had used clean white cotton fabric, so she chose to use white cotton fabric with dirt and a heavier material, red felt, to test the M-Vac™, using a pre-filter before the normal procedure.
University of California-Davis student Vianca Castro-Harris studied several aspects of the M-Vac™ System use in her masters program project, including the efficacy of pre-filtering. Most of the previous studies she reviewed had used clean white cotton fabric, so she chose to use white cotton fabric with dirt and a heavier material, red felt, to test the M-Vac™, using a pre-filter before the normal procedure.
Objective: "The purpose of this study is to provide crime labs with information on the efficacy of the M-Vac™ collection system with pre-filtration on total DNA yield and purity..." Because "evidence items are often contaminated with dirt or other substances including numerous dyes, ... this study was designed to evaluate the effectiveness of the M-Vac™ on a variety of cloth samples including those with dirt present."
Methods: The cotton and felt materials were purchased, washed, cut into swatches, and stored in manila envelopes. Saliva was collected and introduced to its appropriate fabrics. Dirt was collected from two sources, exposed to UV light for an hour to kill any existing DNA, then applied to the white cotton. Fabric swatches were secured in embroidery hoops to make the M-Vac™ sampling easier. The M-Vac™ sampling was performed, with the head moved through a specific collection pattern on each swatch. The liquid collected was split into two parts. "The first half (35ml) was Nalgene (0.45µm M-Vac™ Concentration Filter)* filtered, while the second half of the solution (35ml) was Steriflip® pre-filtered (40µm M-Vac™ Pre-Filter and Pre-Filter Shield)* and then Nalgene filtered." After both kinds of filtering, the Nalgene filter was removed, cut into strips and placed in Lysep tubes.
Extraction: "Samples were retrieved from the freezer and lysed using DTT and Prepfiler™ Lysis buffer.* The DNA was extracted using a Prepfiler™ Kit and the Hamilton STARlet™* liquid handling robot.... After extraction, the samples were stored in the laboratory refrigerator until they were needed for DNA quantification."
Quantification: "The Life Technologies Quantifiler™ Trio Quantification Kit* was used with the Applied Biosystems 7500 Real-Time PCR Instrument,* with various primers."
Amplification: "The GlobalFiler® kit* is a short tandem repeat (STR) multiplex assay that amplifies 21 autosomal STR loci, 1 Y-STR, 1 Yindel and the Amelogenin sex-determining marker in a single PCR amplification.* The Hamilton STARlet™ Robotic platform and GlobalFiler® PCR amplification kit was used for normalization and amplification set-up of a select number of the extracted DNA samples. Of the selected DNA samples, DNA samples with ≤0.05 ng/µL of small autosomal DNA were concentrated in Nanosep® columns* before they were amplified using the ProFlex™ PCR System thermal cycler.* Concentrated samples were stored in the laboratory refrigerator (See DNA concentration protocol in Appendix IV). Samples that had undetermined values for all three quant target regions (large autosomal, small autosomal, and male) were not amplified. The GlobalFiler® protocol is included in Appendix V."
Results and Conclusion: "The single-filtered felt samples yielded 1.13 times more DNA on average than the double-filtered felt samples." In fact, surprisingly, the single-filtered felt samples yielded more DNA, more alleles, and more complete DNA profiles than the double-filtered felt samples. Ms. Castro-Harris had hypothesized that prefiltration would result in significantly higher DNA yields by removing small fabric fibers and dye particles, which are known to inhibit downstream DNA amplification, from the collected M-Vac™ fluid. However, double-filtered felt samples did not have a higher DNA yield on average. Small felt fibers may have trapped some of the biological material onto the first filter, preventing some cellular material from making it through to the filtrate tube. Other possible explanations were also proposed. Though some of the double-filtered red felt samples did exhibit an increased DNA yield, considering the overall results Ms. Castro-Harris says, "We conclude that the Steriflip® pre-filter is not an effective tool to filter inhibitors from red-felt samples." It is important to note the findings were limited by the sample fabric type, the source of DNA (saliva), and the sensitivity of the DNA technology used.
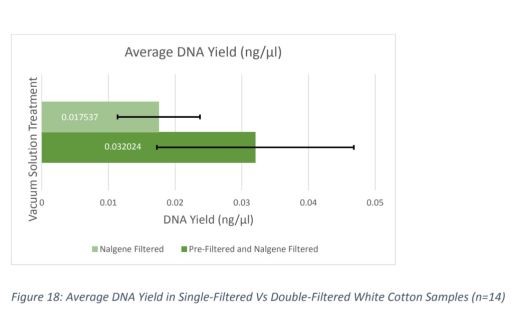 The "double-filtered white-cotton samples with dirt had 1.82 times more DNA on average than single-filtered white-cotton samples with dirt," which was determined not to be "statistically significant under the conditions." "Therefore, results regarding the effectiveness of the M-Vac™ and Steriflip® pre-filter on white-cotton samples with dirt were inconclusive." Again, several options for the cause were suggested. Ms. Castro-Harris' conclusion here is that, "Due to the results of double-filtered cotton samples with dirt, we conclude that a Steriflip® Pre-filter is only a partially effective tool to filter inhibitors from dirt samples."
The "double-filtered white-cotton samples with dirt had 1.82 times more DNA on average than single-filtered white-cotton samples with dirt," which was determined not to be "statistically significant under the conditions." "Therefore, results regarding the effectiveness of the M-Vac™ and Steriflip® pre-filter on white-cotton samples with dirt were inconclusive." Again, several options for the cause were suggested. Ms. Castro-Harris' conclusion here is that, "Due to the results of double-filtered cotton samples with dirt, we conclude that a Steriflip® Pre-filter is only a partially effective tool to filter inhibitors from dirt samples."
*Details about kit titles and sources were omitted for easier reading, but can be made available upon request.
References:Castro, Vianca. Effectiveness of Steriflip® Pre-filters for Removing PCR Inhibitors from Forensic DNA Samples Obtained Using the M-Vac™ Collection System.
Contact us for further details.